|
以决策树作为开始,因为简单,而且也比较容易用到,当前的boosting或random
forest也是常以其为基础的
决策树算法本身参考之前的blog,其实就是贪婪算法,每次切分使得数据变得最为有序
那么如何来定义有序或无序?
无序,node impurity

对于分类问题,我们可以用熵entropy或Gini来表示信息的无序程度
对于回归问题,我们用方差Variance来表示无序程度,方差越大,说明数据间差异越大
information gain
用于表示,由父节点划分后得到子节点,所带来的impurity的下降,即有序性的增益
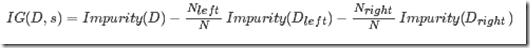
MLib决策树的例子
下面直接看个regression的例子,分类的case,差不多,
import org.apache.spark.mllib.tree.DecisionTree
import org.apache.spark.mllib.util.MLUtils
// Load and parse the data file.
// Cache the data since we will use it again to compute training error.
val data = MLUtils.loadLibSVMFile(sc, "data/mllib/sample_libsvm_data.txt").cache()
// Train a DecisionTree model.
// Empty categoricalFeaturesInfo indicates all features are continuous.
val categoricalFeaturesInfo = Map[Int, Int]()
val impurity = "variance"
val maxDepth = 5
val maxBins = 100
val model = DecisionTree.trainRegressor(data, categoricalFeaturesInfo, impurity,
maxDepth, maxBins)
// Evaluate model on training instances and compute training error
val labelsAndPredictions = data.map { point =>
val prediction = model.predict(point.features)
(point.label, prediction)
}
val trainMSE = labelsAndPredictions.map{ case(v, p) => math.pow((v - p), 2)}.mean()
println("Training Mean Squared Error = " + trainMSE)
println("Learned regression tree model:\n" + model) |
还是比较简单的,由于是回归,所以impurity的定义为variance
maxDepth,最大树深,设为5
maxBins,最大的划分数
先理解什么是bin,决策树的算法就是对feature的取值不断的进行划分
对于离散的feature,比较简单,如果有m个值,最多 个划分,如果值是有序的,那么就最多m-1个划分 个划分,如果值是有序的,那么就最多m-1个划分
比如年龄feature,有老,中,少3个值,如果无序有 个,即3种划分,老|中,少;老,中|少;老,少|中 个,即3种划分,老|中,少;老,中|少;老,少|中
但如果是有序的,即按老,中,少的序,那么只有m-1个,即2种划分,老|中,少;老,中|少
对于连续的feature,其实就是进行范围划分,而划分的点就是split,划分出的区间就是bin
对于连续feature,理论上划分点是无数的,但是出于效率我们总要选取合适的划分点
有个比较常用的方法是取出训练集中该feature出现过的值作为划分点,
但对于分布式数据,取出所有的值进行排序也比较费资源,所以可以采取sample的方式
源码分析
首先调用,DecisionTree.trainRegressor,类似调用静态函数(object DecisionTree)
org.apache.spark.mllib.tree.DecisionTree.scala
/**
* Method to train a decision tree model for regression.
*
* @param input Training dataset: RDD of [[org.apache.spark.mllib.regression.LabeledPoint]].
* Labels are real numbers.
* @param categoricalFeaturesInfo Map storing arity of categorical features.
* E.g., an entry (n -> k) indicates that feature n is categorical
* with k categories indexed from 0: {0, 1, ..., k-1}.
* @param impurity Criterion used for information gain calculation.
* Supported values: "variance".
* @param maxDepth Maximum depth of the tree.
* E.g., depth 0 means 1 leaf node; depth 1 means 1 internal node + 2 leaf nodes.
* (suggested value: 5)
* @param maxBins maximum number of bins used for splitting features
* (suggested value: 32)
* @return DecisionTreeModel that can be used for prediction
*/
def trainRegressor(
input: RDD[LabeledPoint],
categoricalFeaturesInfo: Map[Int, Int],
impurity: String,
maxDepth: Int,
maxBins: Int): DecisionTreeModel = {
val impurityType = Impurities.fromString(impurity)
train(input, Regression, impurityType, maxDepth, 0, maxBins, Sort, categoricalFeaturesInfo)
} |
调用静态函数train
def train(
input: RDD[LabeledPoint],
algo: Algo,
impurity: Impurity,
maxDepth: Int,
numClassesForClassification: Int,
maxBins: Int,
quantileCalculationStrategy: QuantileStrategy,
categoricalFeaturesInfo: Map[Int,Int]): DecisionTreeModel = {
val strategy = new Strategy(algo, impurity, maxDepth, numClassesForClassification, maxBins,
quantileCalculationStrategy, categoricalFeaturesInfo)
new DecisionTree(strategy).train(input)
} |
可以看到将所有参数封装到Strategy类,然后初始化DecisionTree类对象,继续调用成员函数train
/**
* :: Experimental ::
* A class which implements a decision tree learning algorithm for classification and regression.
* It supports both continuous and categorical features.
* @param strategy The configuration parameters for the tree algorithm which specify the type
* of algorithm (classification, regression, etc.), feature type (continuous,
* categorical), depth of the tree, quantile calculation strategy, etc.
*/
@Experimental
class DecisionTree (private val strategy: Strategy) extends Serializable with Logging {
strategy.assertValid()
/**
* Method to train a decision tree model over an RDD
* @param input Training data: RDD of [[org.apache.spark.mllib.regression.LabeledPoint]]
* @return DecisionTreeModel that can be used for prediction
*/
def train(input: RDD[LabeledPoint]): DecisionTreeModel = {
// Note: random seed will not be used since numTrees = 1.
val rf = new RandomForest(strategy, numTrees = 1, featureSubsetStrategy = "all", seed = 0)
val rfModel = rf.train(input)
rfModel.trees(0)
}
} |
可以看到,这里DecisionTree的设计是基于RandomForest的特例,即单颗树的RandomForest
所以调用RandomForest.train(),最终因为只有一棵树,所以取trees(0)
org.apache.spark.mllib.tree.RandomForest.scala
重点看下,RandomForest里面的train做了什么?
/**
* Method to train a decision tree model over an RDD
* @param input Training data: RDD of [[org.apache.spark.mllib.regression.LabeledPoint]]
* @return RandomForestModel that can be used for prediction
*/
def train(input: RDD[LabeledPoint]): RandomForestModel = {
//1. metadata
val retaggedInput = input.retag(classOf[LabeledPoint])
val metadata =
DecisionTreeMetadata.buildMetadata(retaggedInput, strategy, numTrees, featureSubsetStrategy)
// 2. Find the splits and the corresponding bins (interval between the splits) using a sample
// of the input data.
val (splits, bins) = DecisionTree.findSplitsBins(retaggedInput, metadata)
// 3. Bin feature values (TreePoint representation).
// Cache input RDD for speedup during multiple passes.
val treeInput = TreePoint.convertToTreeRDD(retaggedInput, bins, metadata)
val baggedInput = if (numTrees > 1) {
BaggedPoint.convertToBaggedRDD(treeInput, numTrees, seed)
} else {
BaggedPoint.convertToBaggedRDDWithoutSampling(treeInput)
}.persist(StorageLevel.MEMORY_AND_DISK)
// set maxDepth and compute memory usage
// depth of the decision tree
// Max memory usage for aggregates
// TODO: Calculate memory usage more precisely.
//........
/*
* The main idea here is to perform group-wise training of the decision tree nodes thus
* reducing the passes over the data from (# nodes) to (# nodes / maxNumberOfNodesPerGroup).
* Each data sample is handled by a particular node (or it reaches a leaf and is not used
* in lower levels).
*/
// FIFO queue of nodes to train: (treeIndex, node)
val nodeQueue = new mutable.Queue[(Int, Node)]()
val rng = new scala.util.Random()
rng.setSeed(seed)
// Allocate and queue root nodes.
val topNodes: Array[Node] = Array.fill[Node](numTrees)(Node.emptyNode(nodeIndex = 1))
Range(0, numTrees).foreach(treeIndex => nodeQueue.enqueue((treeIndex, topNodes(treeIndex))))
while (nodeQueue.nonEmpty) {
// Collect some nodes to split, and choose features for each node (if subsampling).
// Each group of nodes may come from one or multiple trees, and at multiple levels.
val (nodesForGroup, treeToNodeToIndexInfo) =
RandomForest.selectNodesToSplit(nodeQueue, maxMemoryUsage, metadata, rng)
// 对decision tree没有意义,nodeQueue只有一个node,不需要选
// 4. Choose node splits, and enqueue new nodes as needed.
DecisionTree.findBestSplits(baggedInput, metadata, topNodes, nodesForGroup,
treeToNodeToIndexInfo, splits, bins, nodeQueue, timer)
}
val trees = topNodes.map(topNode => new DecisionTreeModel(topNode, strategy.algo))
RandomForestModel.build(trees)
} |
1. DecisionTreeMetadata.buildMetadata
org.apache.spark.mllib.tree.impl.DecisionTreeMetadata.scala
这里生成一些后面需要用到的metadata
最关键的是计算每个feature的bins和splits的数目,
计算bins的数目
//bins数目最大不能超过训练集中样本的size
val maxPossibleBins = math.min(strategy.maxBins, numExamples).toInt
//设置默认值
val numBins = Array.fill[Int](numFeatures)(maxPossibleBins)
if (numClasses > 2) {
// Multiclass classification
val maxCategoriesForUnorderedFeature =
((math.log(maxPossibleBins / 2 + 1) / math.log(2.0)) + 1).floor.toInt
strategy.categoricalFeaturesInfo.foreach { case (featureIndex, numCategories) =>
// Decide if some categorical features should be treated as unordered features,
// which require 2 * ((1 << numCategories - 1) - 1) bins.
// We do this check with log values to prevent overflows in case numCategories is large.
// The next check is equivalent to: 2 * ((1 << numCategories - 1) - 1) <= maxBins
if (numCategories <= maxCategoriesForUnorderedFeature) {
unorderedFeatures.add(featureIndex)
numBins(featureIndex) = numUnorderedBins(numCategories)
} else {
numBins(featureIndex) = numCategories
}
}
} else {
// Binary classification or regression
strategy.categoricalFeaturesInfo.foreach { case (featureIndex, numCategories) =>
numBins(featureIndex) = numCategories
}
}
|
其他case,bins数目等于feature的numCategories
对于unordered情况,比较特殊,
/**
* Given the arity of a categorical feature (arity = number of categories),
* return the number of bins for the feature if it is to be treated as an unordered feature.
* There is 1 split for every partitioning of categories into 2 disjoint, non-empty sets;
* there are math.pow(2, arity - 1) - 1 such splits.
* Each split has 2 corresponding bins.
*/
def numUnorderedBins(arity: Int): Int = 2 * ((1 << arity - 1) - 1) |
根据bins数目,计算splits
/**
* Number of splits for the given feature.
* For unordered features, there are 2 bins per split.
* For ordered features, there is 1 more bin than split.
*/
def numSplits(featureIndex: Int): Int = if (isUnordered(featureIndex)) {
numBins(featureIndex) >> 1
} else {
numBins(featureIndex) - 1
} |
2. DecisionTree.findSplitsBins
首先找出每个feature上可能出现的splits和相应的bins,这是后续算法的基础
这里的注释解释了上面如何计算splits和bins数目的算法
a,对于连续数据,对于一个feature,splits = numBins
- 1;上面也说了对于连续值,其实splits可以无限的,如何找到numBins - 1个splits,很简单,这里用sample
b,对于离散数据,两个case
b.1, 无序的feature,用于low-arity(参数较少)的multiclass分类,这种case下划分的可能性比较多,image,所以用subsets
of categories来作为划分
b.2, 有序的feature,用于regression,二元分类,或high-arity的多元分类,这种case下划分的可能比较少,m-1,所以用每个category作为划分
/**
* Returns splits and bins for decision tree calculation.
* Continuous and categorical features are handled differently.
*
* Continuous features:
* For each feature, there are numBins - 1 possible splits representing the possible binary
* decisions at each node in the tree.
* This finds locations (feature values) for splits using a subsample of the data.
*
* Categorical features:
* For each feature, there is 1 bin per split.
* Splits and bins are handled in 2 ways:
* (a) "unordered features"
* For multiclass classification with a low-arity feature
* (i.e., if isMulticlass && isSpaceSufficientForAllCategoricalSplits),
* the feature is split based on subsets of categories.
* (b) "ordered features"
* For regression and binary classification,
* and for multiclass classification with a high-arity feature,
* there is one bin per category.
*
* @param input Training data: RDD of [[org.apache.spark.mllib.regression.LabeledPoint]]
* @param metadata Learning and dataset metadata
* @return A tuple of (splits, bins).
* Splits is an Array of [[org.apache.spark.mllib.tree.model.Split]]
* of size (numFeatures, numSplits).
* Bins is an Array of [[org.apache.spark.mllib.tree.model.Bin]]
* of size (numFeatures, numBins).
*/
protected[tree] def findSplitsBins(
input: RDD[LabeledPoint],
metadata: DecisionTreeMetadata): (Array[Array[Split]], Array[Array[Bin]]) = {
val numFeatures = metadata.numFeatures
// Sample the input only if there are continuous features.
val hasContinuousFeatures = Range(0, numFeatures).exists(metadata.isContinuous)
val sampledInput = if (hasContinuousFeatures) { // 对于连续特征,取值会比较多,需要做抽样
// Calculate the number of samples for approximate quantile calculation.
val requiredSamples = math.max(metadata.maxBins * metadata.maxBins, 10000) // 抽样数要远大于桶数
val fraction = if (requiredSamples < metadata.numExamples) { // 设置抽样比例
requiredSamples.toDouble / metadata.numExamples
} else {
1.0
}
input.sample(withReplacement = false, fraction, new XORShiftRandom().nextInt()).collect()
} else {
new Array[LabeledPoint](0)
}
metadata.quantileStrategy match {
case Sort =>
val splits = new Array[Array[Split]](numFeatures) // 初始化splits和bins
val bins = new Array[Array[Bin]](numFeatures)
// Find all splits.
// Iterate over all features.
var featureIndex = 0
while (featureIndex < numFeatures) { // 遍历所有的feature
val numSplits = metadata.numSplits(featureIndex) // 取出前面算出的splits和bins的数目
val numBins = metadata.numBins(featureIndex)
if (metadata.isContinuous(featureIndex)) { // 对于连续的feature
val numSamples = sampledInput.length
splits(featureIndex) = new Array[Split](numSplits)
bins(featureIndex) = new Array[Bin](numBins)
val featureSamples = sampledInput.map(lp => lp.features(featureIndex)).sorted
// 从sampledInput里面取出该feature的所有取值,排序
val stride: Double = numSamples.toDouble / metadata.numBins(featureIndex)
// 取样数/桶数,决定split(划分)的步长
logDebug("stride = " + stride)
for (splitIndex <- 0 until numSplits) { // 开始划分
val sampleIndex = splitIndex * stride.toInt // 划分数×步长,得到划分所用的sample的index
// Set threshold halfway in between 2 samples.
val threshold = (featureSamples(sampleIndex) + featureSamples(sampleIndex + 1)) / 2.0
// 划分点选取在前后两个sample的均值
splits(featureIndex)(splitIndex) =
new Split(featureIndex, threshold, Continuous, List()) // 创建Split对象
}
bins(featureIndex)(0) = new Bin(new DummyLowSplit(featureIndex, Continuous),
// 初始化第一个split,DummyLowSplit,取值是Double.MinValue
splits(featureIndex)(0), Continuous, Double.MinValue)
for (splitIndex <- 1 until numSplits) { // 创建所有的bins
bins(featureIndex)(splitIndex) =
new Bin(splits(featureIndex)(splitIndex - 1), splits(featureIndex)(splitIndex),
Continuous, Double.MinValue)
}
bins(featureIndex)(numSplits) = new Bin(splits(featureIndex)(numSplits - 1),
// 初始化最后一个split,DummyHighSplit,取值是Double.MaxValue
new DummyHighSplit(featureIndex, Continuous), Continuous, Double.MinValue)
} else { // 对于分类的feature
// Categorical feature
val featureArity = metadata.featureArity(featureIndex) // 离散特征中的取值个数
if (metadata.isUnordered(featureIndex)) { // 无序的离散特征
// TODO: The second half of the bins are unused. Actually, we could just use
// splits and not build bins for unordered features. That should be part of
// a later PR since it will require changing other code (using splits instead
// of bins in a few places).
// Unordered features
// 2^(maxFeatureValue - 1) - 1 combinations
splits(featureIndex) = new Array[Split](numSplits)
bins(featureIndex) = new Array[Bin](numBins)
var splitIndex = 0
while (splitIndex < numSplits) {
val categories: List[Double] =
extractMultiClassCategories(splitIndex + 1, featureArity)
splits(featureIndex)(splitIndex) =
new Split(featureIndex, Double.MinValue, Categorical, categories)
bins(featureIndex)(splitIndex) = {
if (splitIndex == 0) {
new Bin(
new DummyCategoricalSplit(featureIndex, Categorical),
splits(featureIndex)(0),
Categorical,
Double.MinValue)
} else {
new Bin(
splits(featureIndex)(splitIndex - 1),
splits(featureIndex)(splitIndex),
Categorical,
Double.MinValue)
}
}
splitIndex += 1
}
} else { // 有序的离散特征,不需要事先算,因为splits就等于featureArity
// Ordered features
// Bins correspond to feature values, so we do not need to compute splits or bins
// beforehand. Splits are constructed as needed during training.
splits(featureIndex) = new Array[Split](0)
bins(featureIndex) = new Array[Bin](0)
}
}
featureIndex += 1
}
(splits, bins)
case MinMax =>
throw new UnsupportedOperationException("minmax not supported yet.")
case ApproxHist =>
throw new UnsupportedOperationException("approximate histogram not supported yet.")
}
} |
3. TreePoint和BaggedPoint
TreePoint是LabeledPoint的内部数据结构,这里需要做转换,
private def labeledPointToTreePoint(
labeledPoint: LabeledPoint,
bins: Array[Array[Bin]],
featureArity: Array[Int],
isUnordered: Array[Boolean]): TreePoint = {
val numFeatures = labeledPoint.features.size
val arr = new Array[Int](numFeatures)
var featureIndex = 0
while (featureIndex < numFeatures) {
arr(featureIndex) = findBin(featureIndex, labeledPoint, featureArity(featureIndex),
isUnordered(featureIndex), bins)
featureIndex += 1
}
new TreePoint(labeledPoint.label, arr) //只是将labeledPoint中的value替换成arr
} |
arr是findBin的结果, 这里主要是针对连续特征做处理,将连续的值通过二分查找转换为相应bin的index
对于离散数据,bin等同于featureValue.toInt
BaggedPoint,由于random forest是比较典型的bagging算法,所以需要对训练集做bootstrap
sample
而对于decision tree是特殊的单根random forest,所以不需要做抽样
BaggedPoint.convertToBaggedRDDWithoutSampling(treeInput)
其实只是做简单的封装
4. DecisionTree.findBestSplits
这段代码写的有点复杂,尤其和randomForest混杂一起
总之,关键在
// find best split for each node
val (split: Split, stats: InformationGainStats, predict: Predict) =
binsToBestSplit(aggStats, splits, featuresForNode, nodes(nodeIndex))
(nodeIndex, (split, stats, predict))
}.collectAsMap() |
看看binsToBestSplit的实现,为了清晰一点,我们只看continuous feature
四个参数:
binAggregates: DTStatsAggregator, 就是ImpurityAggregator,给出如果算出impurity的逻辑
splits: Array[Array[Split]], feature对应的splits
featuresForNode: Option[Array[Int]],
tree node对应的feature
node: Node, 哪个tree node
返回值:(Split, InformationGainStats, Predict),
Split,最优的split对象(包含featureindex和splitindex)
InformationGainStats,该split产生的Gain对象,表明产生多少增益,多大程度降低impurity
Predict,该节点的预测值,对于连续feature就是平均值,看后面的分析
private def binsToBestSplit(
binAggregates: DTStatsAggregator,
splits: Array[Array[Split]],
featuresForNode: Option[Array[Int]],
node: Node): (Split, InformationGainStats, Predict) = {
// For each (feature, split), calculate the gain, and select the best (feature, split).
val (bestSplit, bestSplitStats) =
Range(0, binAggregates.metadata.numFeaturesPerNode).map { featureIndexIdx => //遍历每个feature
//......取出feature对应的splits
// Find best split.
val (bestFeatureSplitIndex, bestFeatureGainStats) =
Range(0, numSplits).map { case splitIdx => //遍历每个splits
val leftChildStats = binAggregates.getImpurityCalculator(nodeFeatureOffset, splitIdx)
val rightChildStats = binAggregates.getImpurityCalculator(nodeFeatureOffset, numSplits)
rightChildStats.subtract(leftChildStats)
predictWithImpurity = Some(predictWithImpurity.getOrElse(
calculatePredictImpurity(leftChildStats, rightChildStats)))
val gainStats = calculateGainForSplit(leftChildStats, //算出gain,InformationGainStats对象
rightChildStats, binAggregates.metadata, predictWithImpurity.get._2)
(splitIdx, gainStats)
}.maxBy(_._2.gain) //找到gain最大的split的index
(splits(featureIndex)(bestFeatureSplitIndex), bestFeatureGainStats)
}
//......省略离散特征的case
}.maxBy(_._2.gain) //找到gain最大的feature的split
(bestSplit, bestSplitStats, predictWithImpurity.get._1)
} |
Predict,这个需要分析一下
predictWithImpurity.get._1,predictWithImpurity元组的第一个元素
calculatePredictImpurity的返回值中的predict
private def calculatePredictImpurity(
leftImpurityCalculator: ImpurityCalculator,
rightImpurityCalculator: ImpurityCalculator): (Predict, Double) = {
val parentNodeAgg = leftImpurityCalculator.copy
parentNodeAgg.add(rightImpurityCalculator)
val predict = calculatePredict(parentNodeAgg)
val impurity = parentNodeAgg.calculate()
(predict, impurity)
} |
private def calculatePredict(impurityCalculator: ImpurityCalculator): Predict = {
val predict = impurityCalculator.predict
val prob = impurityCalculator.prob(predict)
new Predict(predict, prob)
} |
这里predict和impurity有什么不同,可以看出
impurity = ImpurityCalculator.calculate()
predict = ImpurityCalculator.predict |
对于连续feature,我们就看Variance的实现,
/**
* Calculate the impurity from the stored sufficient statistics.
*/
def calculate(): Double = Variance.calculate(stats(0), stats(1), stats(2)) |
@DeveloperApi
override def calculate(count: Double, sum: Double, sumSquares: Double): Double = {
if (count == 0) {
return 0
}
val squaredLoss = sumSquares - (sum * sum) / count
squaredLoss / count
} |
从calculate的实现可以看到,impurity求的就是方差, 不是标准差(均方差)
/**
* Prediction which should be made based on the sufficient statistics.
*/
def predict: Double = if (count == 0) {
0
} else {
stats(1) / count
} |
而predict求的就是平均值
|